discretize.TreeMesh.closest_points_index#
- TreeMesh.closest_points_index(locations, grid_loc='CC', discard=False)[source]#
Find the indicies for the nearest grid location for a set of points.
- Parameters:
- locations(
n,dim)numpy.ndarray Points to query.
- grid_loc{‘CC’, ‘N’, ‘Fx’, ‘Fy’, ‘Fz’, ‘Ex’, ‘Ex’, ‘Ey’, ‘Ez’}
Specifies the grid on which points are being moved to.
- discardbool,
optional Whether to discard the intenally created scipy.spatial.KDTree.
- locations(
- Returns:
- (
n)numpy.ndarrayofint Vector of length n containing the indicies for the closest respective cell center, node, face or edge.
- (
Examples
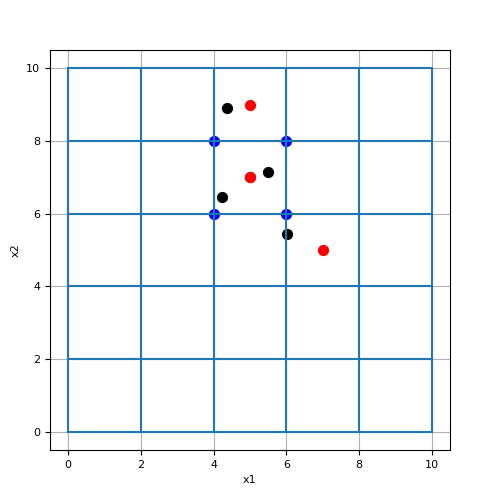
Here we define a set of random (x, y) locations and find the closest cell centers and nodes on a mesh.
>>> from discretize import TensorMesh >>> from discretize.utils import closest_points_index >>> import numpy as np >>> import matplotlib.pyplot as plt >>> h = 2*np.ones(5) >>> mesh = TensorMesh([h, h], x0='00')
Define some random locations, grid cell centers and grid nodes,
>>> xy_random = np.random.uniform(0, 10, size=(4,2)) >>> xy_centers = mesh.cell_centers >>> xy_nodes = mesh.nodes
Find indicies of closest cell centers and nodes,
>>> ind_centers = mesh.closest_points_index(xy_random, 'cell_centers') >>> ind_nodes = mesh.closest_points_index(xy_random, 'nodes')
Plot closest cell centers and nodes
>>> fig = plt.figure(figsize=(5, 5)) >>> ax = fig.add_axes([0.1, 0.1, 0.8, 0.8]) >>> mesh.plot_grid(ax=ax) >>> ax.scatter(xy_random[:, 0], xy_random[:, 1], 50, 'k') >>> ax.scatter(xy_centers[ind_centers, 0], xy_centers[ind_centers, 1], 50, 'r') >>> ax.scatter(xy_nodes[ind_nodes, 0], xy_nodes[ind_nodes, 1], 50, 'b') >>> plt.show()
(
Source code,png,pdf)