discretize.operators.DiffOperators.average_node_to_cell#
- property DiffOperators.average_node_to_cell#
Averaging operator from nodes to cell centers (scalar quantities).
This property constructs a 2nd order averaging operator that maps scalar quantities from nodes to cell centers. This averaging operator is used when a discrete scalar quantity defined on mesh nodes must be projected to cell centers. Once constructed, the operator is stored permanently as a property of the mesh. See notes.
- Returns:
- (
n_cells,n_nodes)scipy.sparse.csr_matrix The scalar averaging operator from nodes to cell centers
- (
Notes
Let \(\boldsymbol{\phi_n}\) be a discrete scalar quantity that lives on mesh nodes. average_node_to_cell constructs a discrete linear operator \(\mathbf{A_{nc}}\) that projects \(\boldsymbol{\phi_f}\) to cell centers, i.e.:
\[\boldsymbol{\phi_c} = \mathbf{A_{nc}} \, \boldsymbol{\phi_n}\]where \(\boldsymbol{\phi_c}\) approximates the value of the scalar quantity at cell centers. For each cell, we are simply averaging the values defined on its nodes. The operation is implemented as a matrix vector product, i.e.:
phi_c = Anc @ phi_n
Examples
Here we compute the values of a scalar function on the nodes. We then create an averaging operator to approximate the function at cell centers. We choose to define a scalar function that is strongly discontinuous in some places to demonstrate how the averaging operator will smooth out discontinuities.
We start by importing the necessary packages and defining a mesh.
>>> from discretize import TensorMesh >>> import numpy as np >>> import matplotlib.pyplot as plt >>> h = np.ones(40) >>> mesh = TensorMesh([h, h], x0="CC")
Then we Create a scalar variable on nodes
>>> phi_n = np.zeros(mesh.nN) >>> xy = mesh.nodes >>> phi_n[(xy[:, 1] > 0)] = 25.0 >>> phi_n[(xy[:, 1] < -10.0) & (xy[:, 0] > -10.0) & (xy[:, 0] < 10.0)] = 50.0
Next, we construct the averaging operator and apply it to the discrete scalar quantity to approximate the value at cell centers.
>>> Anc = mesh.average_node_to_cell >>> phi_c = Anc @ phi_n
Plot the results,
>>> fig = plt.figure(figsize=(11, 5)) >>> ax1 = fig.add_subplot(121) >>> mesh.plot_image(phi_n, ax=ax1, v_type="N") >>> ax1.set_title("Variable at nodes", fontsize=16) >>> ax2 = fig.add_subplot(122) >>> mesh.plot_image(phi_c, ax=ax2, v_type="CC") >>> ax2.set_title("Averaged to cell centers", fontsize=16) >>> plt.show()
(
Source code,png,pdf)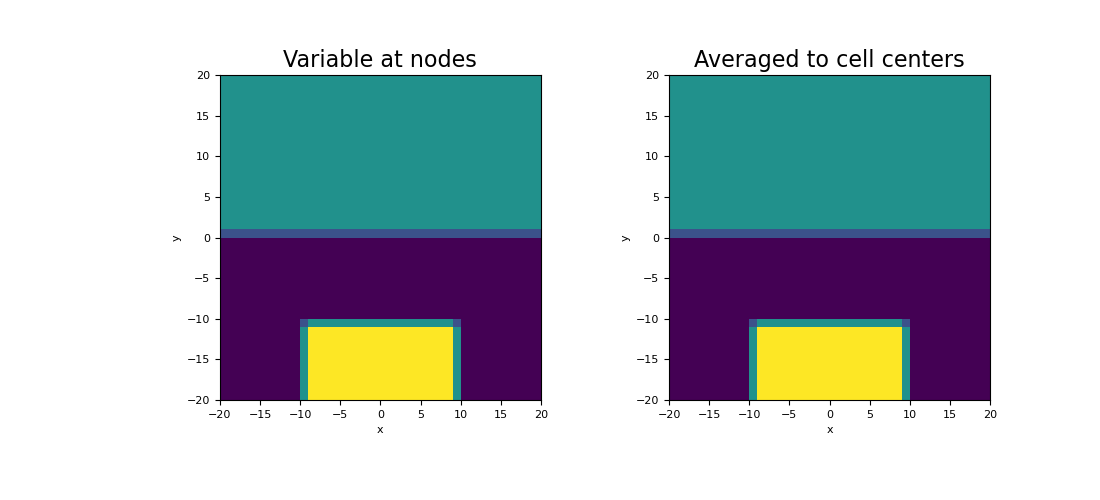
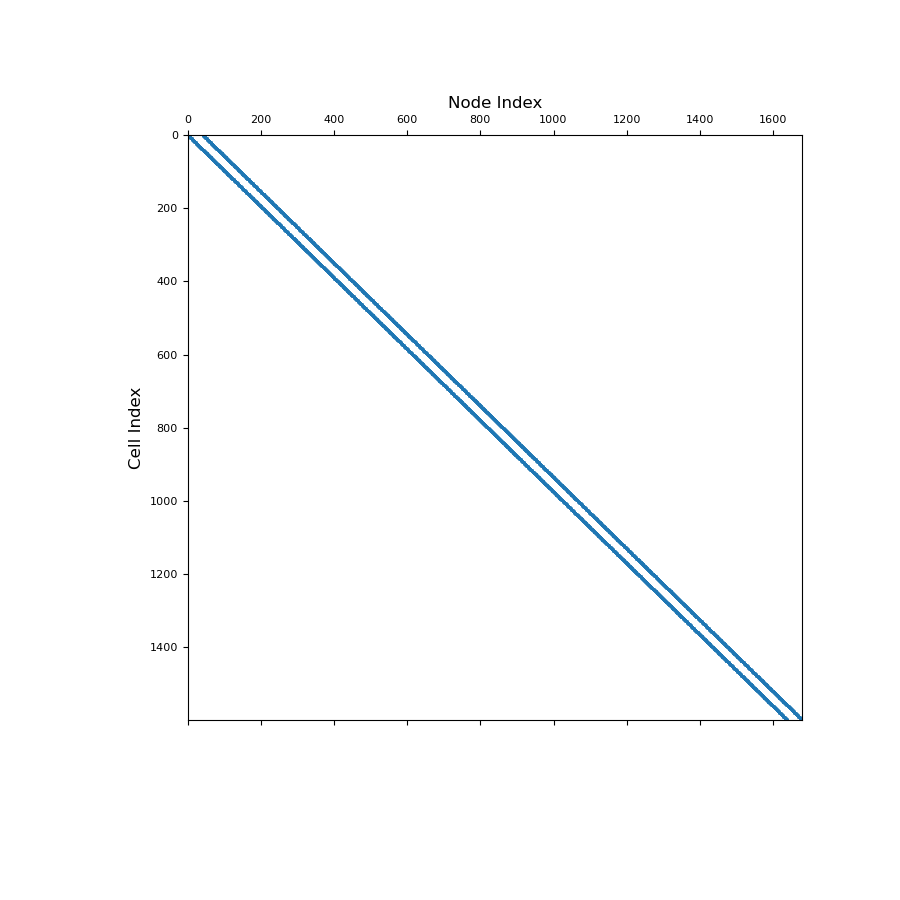
Below, we show a spy plot illustrating the sparsity and mapping of the operator
>>> fig = plt.figure(figsize=(9, 9)) >>> ax1 = fig.add_subplot(111) >>> ax1.spy(Anc, ms=1) >>> ax1.set_title("Node Index", fontsize=12, pad=5) >>> ax1.set_ylabel("Cell Index", fontsize=12) >>> plt.show()